Constructing phenotype algorithms (PAs) is a primary method for both defining diseases and identifying subjects at risk for disease in observational research. While the role of PAs is crucial for effective, reproducible research, the ability to complete detailed PA evaluations has traditionally been limited due to both cost and efficiency.
Lead author Joel Swerdel provided a potential solution to this challenge in PheValuator: Development and evaluation of a phenotype algorithm evaluator, published in the latest issue of the Journal of Biomedical Informatics. Utilizing tools within the OHDSI Network, the research team developed a method that showed promise for reliable phenotype evaluation without reliance on manual review of patient data.
The results were promising for the future use of PheValuator as a means to efficiently evaluate phenotype algorithms. The research demonstrated the use of the tool on four phenotypes, two acute phenotypes, myocardial infarction and cerebral infarction, and two chronic phenotypes, atrial fibrillation and chronic kidney disease. They evaluated four PAs for each phenotype on five databases, four insurance claims databases and one electronic health record database. (Specific tables for each of the four phenotypes are available in the paper.)
Evaluation of such performance characteristics of PAs would have invaluable benefits to researchers. The tool would also allow researchers to compare various phenotype algorithms and select one with appropriate characteristics for a particular study.
The PheValuator results also compared favorably to results from previous PA validation studies across the four phenotypes.
 “We think the use of PheValuator can significantly improve the way we do observational research,” Swerdel said. “We’ll no longer have to guess if we are using the right phenotype algorithm for our work. Eventually we think the output from the tool will be used to allow us to account for and then remove some of the bias that is inherent in observational studies, thereby improving the overall quality and reproducibility of these studies.”
“We think the use of PheValuator can significantly improve the way we do observational research,” Swerdel said. “We’ll no longer have to guess if we are using the right phenotype algorithm for our work. Eventually we think the output from the tool will be used to allow us to account for and then remove some of the bias that is inherent in observational studies, thereby improving the overall quality and reproducibility of these studies.”
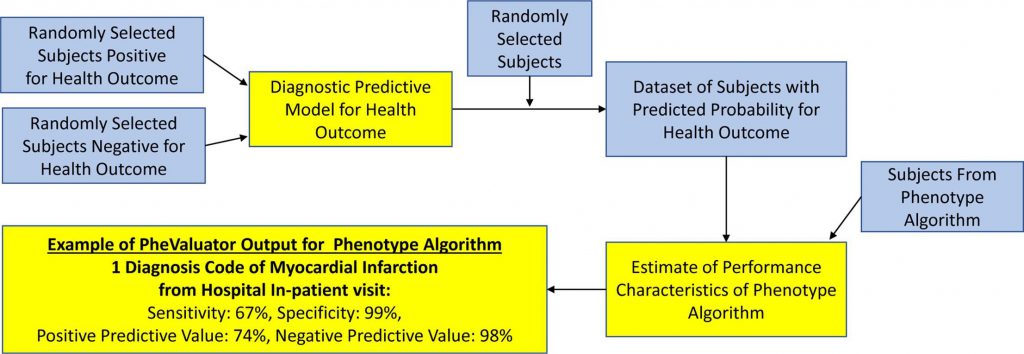
Developing PheValuator (which is available for implementation through GitHub) required the development of a predictive model for a phenotype, which was done through utilization of the OHDSI PatientLevelPrediction (PLP) R package. Following the creation of extremely specific (“xSpec”) and sensitive cohorts, the researchers formed a diagnostic prediction model that helped determine the probability of a phenotype for each individual within a large group.
Testing PheValuator required the conversion of five significant datasets to the OMOP common data model (CDM), which standardizes both structure and content of the observational data. The four-step process for ensuring data quality in the conversion is available within the paper.
The research team notes that future work can both improve the predictive modeling approach and further validate the approach using additional phenotypes and data sources. The current results, however, indicate a potentially useful tool for future observational research.

