The first COVID-19 prediction model developed and validated by the OHDSI community following the March 2020 global study-a-thon was recently published by BMC Medical Research Methodology.
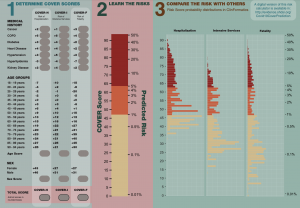
The study “Seek COVER: using a disease proxy to rapidly develop and validate a personalized risk calculator for COVID-19 outcomes in an international network” developed COVID-19 Estimated Risk (COVER) scores that quantify a patient’s risk of hospital admission with pneumonia (COVER-H), hospitalization with pneumonia requiring intensive services or death (COVER-I), or fatality (COVER-F) in the 30-days following COVID-19 diagnosis using historical data from patients with influenza or flu-like symptoms and tested this in COVID-19 patients.
Led by co-first authors Ross Williams and Aniek Markus, both of whom are PhD students at Erasmus University Medical Center, the team designed a nine-predictor risk model that was validated using more than 44,500 COVID patients (following initial development and validation using more than 6.8 million patients with influenza or flu-like symptoms). This model predicts hospitalization, intensive services, and death, and can help provide reassurance for low-risk patients, while shielding high-risk patients, as many start to enter the de-confinement stage of the pandemic.
The team of 49 co-authors helped design this prediction model which is available as an interactive app at: http://evidence.ohdsi.org/Covid19CoverPrediction. The model was first posted on MedRxiv in late May, when there was limited data available globally on COVID-19. The OHDSI community was able to access some of the earliest data thanks to global cooperation, especially from collaborators in South Korea, and the foundation of open science that the community has built upon. You can learn more about the study-a-thon here.
Both Williams and Markus shared some thoughts about this research following its publication:
 1) How were you able to develop a prediction model so early in the pandemic with such little data?
1) How were you able to develop a prediction model so early in the pandemic with such little data?
The amount of data needed to evaluate model performance reliably is much less than the amount needed to train a model. Early on in the pandemic we quickly reached the level needed for model evaluation, but model development would have been more problematic. Therefore, we decided to use a proxy disease (influenza) to preserve the COVID-19 data that we had available. Our assumption was that the people vulnerable to influenza would have similar characteristics as those vulnerable to COVID-19. The large amount of historic influenza cases allowed us to overcome the issues of model development with small data samples. After model training we evaluated the model on data from COVID-19 patients to evaluate model performance reliably.
2) When the model was shared via preprint, are you aware of how it was used and what impact it had?
The COVER scores were used for strategic planning purposes by hospitals and regional governments as well as for risk assessment purposes by institutions planning their office work strategies.
3) How does this work better prepare us for similar disease situations in the future?
This work has two implications for future strategies. First, our work shows the use of a proxy disease can be a fast and effective way to produce a model when data is scarce and time limited (e.g. in case of a new pandemic). Second, it implies that rare diseases could also be targeted for prediction modeling with a similar strategy to overcome the lack of data.
4) How did OHDSI tools and the open nature of the community make work like this possible (or at least more manageable)?
The OHDSI tools and community were an invaluable resource in the production of this model; without the standardisations of the OMOP CDM this work encompassing data from 14 sources and 6 countries would not have been possible. This combined with existing standardised tools meant that the technical elements could flow seamlessly. Furthermore, the open and collaborative nature of the OHDSI community allowed for the rapid progression of experimental design; multiple teams worked simultaneously on the various constituent parts, and as such as soon as decisions were made on, for example, what target cohort to use there was already a definition developed by the phenotyping team ready to be used.


